Research
Metabolic genes take up a huge fraction of a cell’s proteome. These genes share great similarity across organisms and tissues. What defines each cell’s unique metabolic program is the activity carried by each gene, i.e. metabolic flux. We are interested in developing quantitative measurement of metabolic flux across different scales, and taking these approaches to understand the plasticity and robustness of metabolic network in systems including yeasts, cancer and immune cells, and microbiome. We hope to gain quantitative knowledge that will guide design for engineering their metabolism.
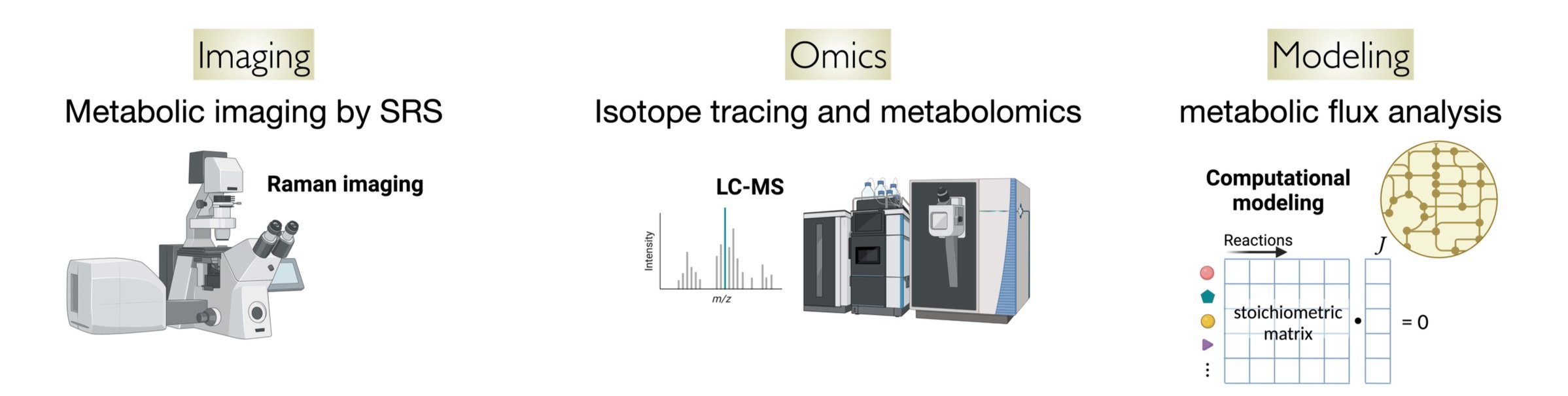
Our science is technique-driven and discovery-driven. We take interdisciplinary approaches from optical imaging and spectroscopy, mass spectrometry, and computational modeling.
Understanding metabolism with greater resolution
By taking advanced approaches in quantitative metabolism, we aim to
develop tools to resolve metabolism in single cells, e.g.
in the tumor microenvironment, or in the gut microbiota
understand quantitative principles governing metabolism, e.g.
How are metabolic programs controlled to ensure robustness, to achieve plasticity, and to support cell function?
How do metabolism of cancer and immune cells adapt to the tumor microenvironment?
How do metabolic interactions shape the composition and structure of microbial communities in the gut?
Imaging metabolism with stimulated Raman scattering (SRS) microscopy
Raman imaging probes the vibration of chemical bonds with light. The frequency shift between the scattered photon and the incident photon matches with the vibrational frequency that is characteristic of a chemical bond. Stimulated Raman scattering microscopy allows fast Raman imaging with high spectral and spatial resolution. By introducing isotopic labels with distinct Raman frequency, we could trace metabolic activity in the cell. Read More
Systems biology of metabolism
For decades we have known that many proliferative cells switch from energy-efficient respiration to energy-inefficient fermentation (Crabtree effect in yeast or Warburg effect in tumor). However, switching energy pathway is only one of the metabolic changes occurring in these cells. How do the entire network rewire to meet the metabolic requirement of proliferation? Do cancer cells and immune cells use different metabolic programs? What is the control logic used to achieve these metabolic adaptations? We address these questions with omics tools based on mass spectrometry and computational tools known as metabolic flux analysis.
We detect hundreds of metabolites (metabolomics) using liquid chromatography-mass spectrometry (LC-MS). Based on these experimental measurements, we infer metabolic flux from genome-reconstructed metabolic models. This has allowed us to resolve metabolic activity through hundreds of reactions in yeasts, cancer cells, and cytotoxic T cells. Read More about how we integrate fluxomics with proteomics to evaluate metabolic efficiency of different pathways.